Publication
808
Phys. Chem. Chem. Phys., 18, 30029-30039, 2016
DOI:10.1039/C6CP05105G
|
|
|
|
|
|
 |
Interaction of osmium(II) redox probe with DNA: insights from theory |
|
|
|
Ashwani Sharma, Sebastien Delile, Mohamed Jabri, Carlo Adamo, Claire Fave, Damien Marchal, and Aurelie Perrier
Chimie ParisTech, PSL Research University, CNRS, Institut de Recherche de Chimie Paris (IRCP), F-75005 Paris, France
Laboratoire d'Electrochimie Moléculaire, UMR 7591 CNRS, Université Paris Diderot, Sorbonne Paris Cité, 15 rue J-A de Baif, F-75205 Paris Cedex 13, France
E-pôle de génoinformatique, Institut Jacques Monod, UMR7592, CNRS, Université Paris Diderot, Sorbonne Paris Cité, F-75013 Paris, France
Institut Universitaire de France, 103 Boulevard Saint Michel, F-75005 Paris, France
Université Paris Diderot, Sorbonne Paris Cité, 5 rue Thomas Mann, F-75205 Paris Cedex 13, France
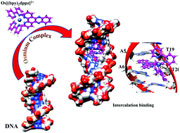
In the course of developing ultrasensitive and quantitative electrochemical point-of-care analytical tools for genetic detection of infectious diseases, osmium(II) metallointercalators were revealed to be suitable and efficient redox probes to monitor the in vitro DNA amplification [Defever etal, Anal. Chem., 2011, 83, 1815–1821]. In this work, we thus propose a complete computational protocol in order to evaluate the affinity between Os(II) complexes with double-stranded DNA. This protocol is based on molecular dynamics, with the parametrization of the GAFF force field for the Os(II) complexes presenting an octahedral environment with polypyridine ligands, and QM/QM′ calculations to evaluate the binding energy. For three Os(II) probes and different binding sites, molecular dynamics simulations and interaction energies calculated at the QM/QM′ level are successively discussed and compared to experimental data in order to identify the most stable binding sites. The computational protocol we propose should then be used to design more efficient Os(II) metallointercalators. |

